Simple COVID-19 Simulator
A downloadable project
Title: Simple COVID-19 Simulator
Platform: MSX
Author: Oscar Urra (Twitter: @o_urra)
Language: MSX-BASIC
Category: SCHAU
PROGRAM DESCRIPTION
This is a small simulator somehow inspired by the classic Conway's Game of life.
In our case, we simulate the propagation of the COVID-19 in a grid, starting
with the following parameters:
- Initial infections: Number of initial cells that are infected, set to 8.
- Death rate: The average % of cells or individuals that shall die. Set to 2 %.
- R0: The Basic Reproduction Number [1], an epidemiological parameter that
measures the expected number of cases directly generated by one case in a
population where all individuals are susceptible to infection. Set to 4.
- Recovery time: Number of days necessary for an infected individual to recover
from the disease. Set to 10.
If we vary these starting parameters, we could also simulate other epidemic
diseases (e.g. Ebola, Influenza, SARS, etc.) since those values can be easily
found and set by modifying the variables in the program (see below).
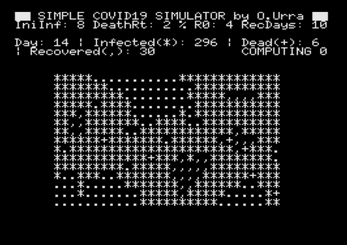
The cells or individuals in the grid can be in any of the states from the SIRD
model [2], that are the following:
- SUSCEPTIBLE, shown in the simulation as a dot '.'. The individual is vulnerable
to the infection and can get the disease.
- INFECTED, shown in the simulation as an asterisk '*'. The individual got the
disease and is infected.
- RECOVERED, shown in the simulation as a comma ','. The individual is recovered
from the disease once the 'recovery days' are elapsed. For the sake of simplicity
we assume that he/she becomes immune and will not be infected again, although
that seems to be not the case with COVID-19.
- DEAD, shown in the simulation as a cross '+'. The individual got infected,
he/she could not recover from the disease, and finally died.
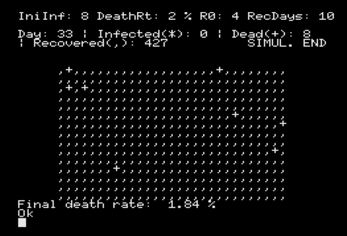
The simulation consists of a number of iterations where the state of every cell
is computed, and it keeps running until the number of infected cells is equal to
zero. Then, the actual death rate is shown and the program ends.
The simulation performance is poor: it's a 8 bit computer after all, and the
status of each of the 435 cells must be computed and updated in every iteration.
Therefore, the program indicates if it is computing the new state (alternating
a 0 and a 1 to show it's not blocked), or if the display is being refreshed. As
an example, a typical simulation can last 39 iterations and take about 23 minutes
to complete, so be patient!
[1] https://en.wikipedia.org/wiki/Basic_reproduction_number
[2] https://en.wikipedia.org/wiki/Mathematical_modelling_of_infectious_disease#The_SIRD_model
STARTING VIA EMULATOR
These instructions are for loading the program in the WebMSX emulator:
1) Open the URL: https://webmsx.org in a modern browser
2) Click the icon of 'Diskette A' (in the lower left corner) and select the
option "Import files to disk"
3) Browse your computer and select the file COVIDSIM.BAS. A diskette with the
file will be 'inserted' in the emulator
4) In the BASIC prompt type: LOAD "COVIDSIM.BAS"
5) Type LIST to see the BASIC code, or RUN to execute the program
6) To stop the program, press CTRL+STOP, which in this the emulator is mapped to
CTRL+F9. With SHIFT+ALT+ARROWS you can speed up the emulation.
PROGRAM STRUCTURE
VARIABLES
H, W: Height and width of the grid.
M%, N%: Arrays with the cells data. The % defines the arrays to contain integers,
which make operating with them a bit faster. M contains the current data and
N contains the temporary data while it's being updated. Every array position
contains and integer with the following meaning:
-2: Recovered cell
-1: Dead cell
0: Uninfected/susceptible cell
1..RT: Infected cell. The integer counts the number of days the cell has
been in that state
F: Number of infected cells
DR: Death Rate in %
R0: The R0 parameter
T: Time, in days, of every iteration of the simulation
D: Counter for the number of dead cells
RC: Counter for the number of recovered cells
RT: Recovery time, in days
Setting different starting values in F, DR, R0 and RT will allow us to simulate
other diseases or conditions.
LINES
10 Clear the screen and initialize the variables and the random numbers generator
20 Print the program name and the initial parameters value
30 Initialize the array with random positions for the initial infected cells.
The positions that are not explicitly initialized are set to 0 by default,
which is the state of 'Uninfected/susceptible'.
40 Beginning of the update loop. For every cell in the array, increment the
days counter and set the cell state to Recovered (-2) if it has been
infected for more than RT days. Also, prints a 0 or 1 depending on whether
the X index is odd or even, to act as an activity indicator.
50 More updates: If the cell is infected, calculate (randomly) the chance that
it dies (state -1). Otherwise, increment the counter of days it's been
infected. Also, if a susceptible cell is surrounded by one or more infected
cells, it also gets infected (randomly) according to R0. In order to avoid
accessing invalid array coordinates, the first and last rows and columns do
not include valid data, and they are not processed
60 Next index values for the update loop
70 Print in the screen the number of cells in every state
80 Print the cells array using a different character for every state
90 If there are still infected cells, go to line 40 and compute another iteration
100 End of simulation. Compute and print the actual death rate and exit| Status | Released |
| Category | Other |
| Author | BASIC 10Liner |
| Tags | 8-Bit, basic, basic10liner, MSX |
Development log
- Image downloadApr 08, 2021
Leave a comment
Log in with itch.io to leave a comment.